Most Read
Abstract
Objective: Hepatic tissues of different patients exposed to systemic bacterial endotoxins are at risk of exposure to so-called lipopolysaccharide (LPS)-unfolded protein response (UPR)- calcium overload-gene splicing (LUCOGS). The essential workings mechanism of LUCOGS and their association with each other to start sepsis-related processes are not elucidated completely. This study aims to unravel the correlation between hepatic gene-splicing and the UPR machinery, in vivo.
Methods: LPS was extracted from the patients’ burned wounds after Pseudomonas aeruginosa isolation (hLPS), with informed consent. The male c57/BL6 mice were divided into six groups: 1. negative control (received sterile pyrogen-free normal saline); 2. positive control received tunicamycin; 3. hLPS (3 mg/kg/intraperitoneal); 4. hLPS+ dantrolene sodium (calcium channel inhibitor, CCI1) (A); 5. hLPS+2-aminomethyl phenyl borinate (1 mg/kg) (CCI2) (B); and 6. combined treatment with hLPS+CCI1/CCI2 (C). Subsequently, mice livers were extracted to evaluate XBP-1 splicing as a marker of the adaptive UPR activation after 2, 8, and 24 hrs.
Results: Compared to the hLPS group, all three CCIs combined with hLPS exhibited a significant increase in the number of XBP-1 splicing. Compared to controls, Group C showed a maximum increase in the number of XBP-1 splicing after 8 and 24 hrs.
Conclusions: Our results indicated that a combination of both CCIs could prevent the pro-inflammatory activation induced by hLPS, via UPR gene-splicing modifications. A new recipe of two types CCIs would potentially prevent the Ca+2 overload intercellular after induced-endotoxemia. Prescription of both CCIs had higher effectiveness than separately to inhibit (pro-) inflammatory processes and could be considered a new strategy to appropriately manage and prevent both LUCOGS in systemic blood circulation and organ shutdown. The effectiveness of the new formula needs complementary studies to further clarify the introduced association mechanism, in vivo.
INTRODUCTION
Different infected patients exposed to systemic bacterial infections of their hepatic tissue are at risk of exposure to acute and severe septic-related processes (SRPs) (1-8). One of the acute side effects is (epi) genetic changes, which aggravate pro-inflammatory responses and increase tissue damage. (3-7). How endotoxins affect gene splicing, and SRPs is not elucidated completely.
In our previous study, we showed that both chemical and wild-type lipopolysaccharide (LPS) could affect TNF-α and IL-1β levels and increase the risk of pro-inflammatory response, which could be inhibited by the appropriate combination of calcium channels inhibitors (CCIs) (8).
In this study, we aim to focus more on details about the so-called LPS-unfolded protein response (UPR)-calcium overload-gene splicing (LUCOGS) working system and its side effects, in vivo (Figure 1 depicts the theoretical model system).
The essential components of the LUCOGS signaling pathway consist of 4 main key phases: 1. Initiating phase; infecting systemic blood circulation, i.e., different effects of lipopolysaccharides (LPS), lipooligosaccharide (LOS); 2. propagating phase; spreading noxious endotoxins metabolites, i.e., reactive oxygen species (ROS) and NO+NO2 (NOX) products, (8); 3. the unfolded protein response (UPR) activation and its disturbance in managing gene splicing and correlated signal transductions (9-14) and; 4. terminating phase; increasing toxic calcium overload (15-22), hyperactivities of hepatocytes (23, 24), and random organ shutdown due to severe intoxication (25-27).
Considerable evidence has revealed that after microbial infections and/or antibiotics cure, LPS is released into the systemic blood circulation and/or the lymphatic system, acts as a stimulator of the immune system in certain patients, which can increase the risk of cell and tissue cytokine release, gene modification, and random organ shutdown (28, 29).
Gram-negative bacterial LPS acts as extremely strong stimulator of innate or natural immunity, which can increase the risk of septic shock and death (1-5, 25-27). The mechanisms of LPS’s induced random organ shutdown and the exact subsequent processes affecting (epi-)genetic changes in hepatic cells are not elucidated completely, in-vivo. In certain patients, after tissue damage and burned-tissue-injuries (30), different infections and inflammations are the main fundamental concerns to treat and/or prevent ultimate consequences. Increased LPS concentrations in chronic infections can prime (pro-)inflammatory responses and SRPs (6-8, 15-20).
One of the most common bacteria causing infection and inflammatory response is Pseudomonas aeruginosa, playing a certain role as a pathogen in a certain fatal infection by its enriched virulence factors (32-35). Primary infections caused by P. aeruginosa have been observed in the respiratory tract, bladder, ears, and ulcers from burns, wounds, and surgical sites. This bacterium is one of the main nosocomial pathogen, which can increase the risk of mortality and morbidity rates in outpatients and inpatients (34).
Different biological basic research revealed that LPS might consist of poly- or oligosaccharide regions, which are anchored in the outer bacterial membrane (1-8).LPS released from attacked and destroyed bacteria induces the migration of neutrophils from the spleen to the location of T-lymphocytes (6, 15, 27).
The liver is another systemic organ playing a prominent antitoxic role against endotoxins and after production of LPS (anti-) inflammatory responses (23, 24). Increasing the concentration of liver enzymes is considered a marker in the acute phase of the inflammation process. Ongoing infections and inflammation might occur in systemic blood circulation after infected tissues are harshly damaged, for example, in patients with severe burns (30, 31).
Different studies have highlighted the lethal role of the Ca+2 ions overloads intracellularly, which might initiate cell death during exposure to endotoxins (9-14). However, to overcome the abovementioned cell survival failure, different strategies were developed by the immune system i.e. Ca+2 ions-related gene expression (36). The shift in gene expression is even found in the patient’s P. aeruginosa microflora, which displays an epidemic population structure (37, 38). Different kinds of gene expression could lead to different outcomes. For example, UPR, in eukaryotic cells like human cells, is a homeostatic response mechanism, which may react to certain endotoxins after exposure to LPS via regulated UPR-inositol requiring enzyme-1(IRE-1) sensors (14-17). This response consecutively protects the subject from different SRPs and/or random organ shutdown. Any kind of endoplasmic reticulum (ER) stressors may initiate unconventional splicing of transcription factor mRNA, i.e., unspliced X-box binding protein-1 (uXBP-1), in the cytoplasm. However, it is proposed that the RNA ligase removes 26-ribonucleotide intron from the cytosolic domain of activated IRE-1 to produce spliced X-box binding protein-1 (sXBP-1). After splicing, the open reading frame shifts transcriptional actions and changes UPR activity outcomes. The uXBP-1 translation leads to unstable protein production. Conversely, Back SH, et al. (39) postulated that sXBP-1 was translated to a stable and functional protein, which could control the transcription of many other correlated UPR genes.
There are two different UPRs that each have their own specific function and activities. The UPR-specific-splicing’s activities (sXBP-1) could be built in a cellular and molecular system, which acts as a cytoprotective (adaptive UPR) or leads to cell death (terminal UPR) (40). Different cell faith can occur after shifting between the abovementioned UPRs forming and sequential activities. For instance, the Ca2+ overload could be a hot point to activate terminal UPR activation that leads to cell death (41).
In this study, we hypothesized that pro-inflammatory activation by LPS could be prevented by using an appropriate combination of CCIs.
METHODS
The whole research study design and materials were divided into two main research: (I) isolation of human-tissue-derived bacteria and subsequent extraction of (wild) LPS (hLPS); and (II) investigation of hLPS-inflammation-induced responses of UPR and XBP-1 splicing. In short, the first research studies were conducted in 5 consecutive steps: 1. Clinical sampling of patients with severely burned wounds; 2. Biochemical identification, confirmation and analysis of isolated bacteria P. aeruginosa (Figure 2); 3. Extraction of hLPS from isolated bacteria and quality control of the extraction method by using HPLC confirmation; 4. Construction of novel study design in-vivo-animal modeling to compare wild-purified hLPS versus chemical standard LPS from Sigma-Aldrich Germany (control), without and with calcium channels inhibitors (CCIs), i.e. Dantrolene sodium CCI1 (Sigma-Aldrich, Germany) (CCI1), 2-Aminoethyl diphenyl borinate (2A-D) (CCI2), and 5. Analysis of molecular biologic experiments and evaluation of changes in the XBP-1 mRNA-gene splicing in mice’s liver.
The Clinical Sampling of Patients with Burned Wounds
The whole procedure was fully explained to the subjects, and after obtaining informed consent, isolation of the samples was carried out. Patients with more than 40% severe burned wounds, and probability of infection with P. aeruginosa infection characteristics were selected. Samples were collected from patients from the Surgery Department of Velayat Hospital Rasht, Rasht, Iran, using sterile swabs and immediately transferred to the Stewart culture medium (Merck, Germany). Swabs were then inoculated to the BHI medium (Merck, Germany), separately cultured on cetrimide (Merck, Germany) and blood agar (Millipore, Germany), in aseptic conditions and finally incubated at 42°C for 48 hours.
Biochemical Identification of Bacteria Isolated from Patients
Grown colonies in the cetrimide and blood agar with specific pigmentation were prepared on the slide for the Gram staining kit (Parsian Teb, Tehran, Iran). Additional biochemical and complementary experiments were conducted to determine bacterial identities, such as citrate, oxidase, indole, urea, and triple sugar iron agar (TSI) (Millipore, Germany).
LPS Extraction
Extraction of endotoxin was carried out by using a specific LPS intron purification kit based on the manufacturer’s instructions. The extracted endotoxin was visualized via 12% polyacrylamide gel electrophoresis (PAGE), stained with silver nitrate, and kept at 4°C until the next use. Then, high-performance liquid chromatography (HPLC) fractionation was used to evaluate and confirm the purity of isolated hLPS versus standard control P. aeroginosa LPS purchased from Sigma, Germany (SIGMA-Cat.no: L8643).
Animal Modeling and Molecular Biologic Assays
Male c57/BL6 mice were randomly selected to be studied (six to eight weeks old/25 ± 1 grams). All animals were equally exposed to 12-12 light-dark circadian cycles and had unrestricted access to water and food during the experiment. The animals were divided into five groups, and each group (n=6) was divided into three-time intervals (2, 8, and 24 hrs.) as follows: 1) Negative control; 2) Positive control; 3) Group, which received CCI1+hLPS; 4) Group, which received CCI2+hLPS, and finally; 5) Group, which received a combination of both calcium channel inhibitors (CCI1+CCI2)+hLPS.
Negative control animals received sterile pyrogen-free normal saline. Animals in the positive controls group were treated with 3 mg/kg/intraperitoneal (IP)-injection of tunicamycin (2.5 mg/kg) to induce ER stress artificially, and the injection of P. aeruginosa LPS was commercially purchased from Sigma, Germany (3 mg/kg), as proposed by Nemzek et al. (42), according to the established toxemia model, and based on previous studies (Figure 4, A). Animals in Group 3 received hLPS (3 mg/kg, IP) +CCI1 (40 mg/kg). Animals in Group 4 received CCI2 (1 mg/kg) + hLPS (3 mg/kg, IP). Finally, the last animal group (group 5) received a combination of CCI1 (40 mg/kg) + CCI2 (1 mg/kg) + (3 mg/kg, IP). In all experimental study groups, the beta-actin house-keeping gene (155 bp in length) was amplified as an internal control of molecular tests.
Molecular Experiments
Along with isolation, verification, quality controls, and immunological assay, the splicing of the XBP-1 mRNA gene was studied in the isolated liver tissue of mice. All surgical instruments were sterilized and were set DNase-RNase free/non-pyrogen for surgical removal and separation of animal liver tissue. Small 10-mg pieces of the isolated liver were immediately frozen after aseptic transfer to the DNase-RNase free/non-pyrogen sterile microtube.
The DNA and RNA of all prepared specimens were extracted using specific kits and according to the manufacturer’s guidelines, and then RT-PCR was performed. Using the spectroscopic technique (NanoDrop-2000c spectrophotometers, Thermo Fisher Scientific, USA), the purity of the extracted RNA was measured. Prior to the cDNA synthesis of extracted RNAs, RNA samples were treated with a DNase specific kit (Sina Clone, Iran) to eliminate possible contamination with DNA. After treatment with a DNase-specific kit, the samples were subjected to RT-PCR (Thermo Fisher Scientific, USA). For PCR, a sensitive and specific primer pair was used for test and control groups. XBP-1 forward primer: 5`GAACCAGGAGTTAAGAACACG3`, and reverse primer: 5`AGGCAACAGTGTCAGAGTCC3` (size of PCR product: unspliced uXBP-1=205 bp and spliced sXBP-1=179 bp) as described (43), and β-actin housekeeping primers were obtained from Cinna gen, Iran.
Statistical Analysis
The statistics of our descriptive results were reported at three-time intervals at 2, 8, and 24 hours after each injection using the number of splicing in animals’ livers compared to the total number of animals. The results of the experiment were analyzed by using the IBM-SPSS software version 16 via χ2 statistic tests at the significant level (p< 0.05). All data were expressed as mean ± standard deviation.
RESULTS
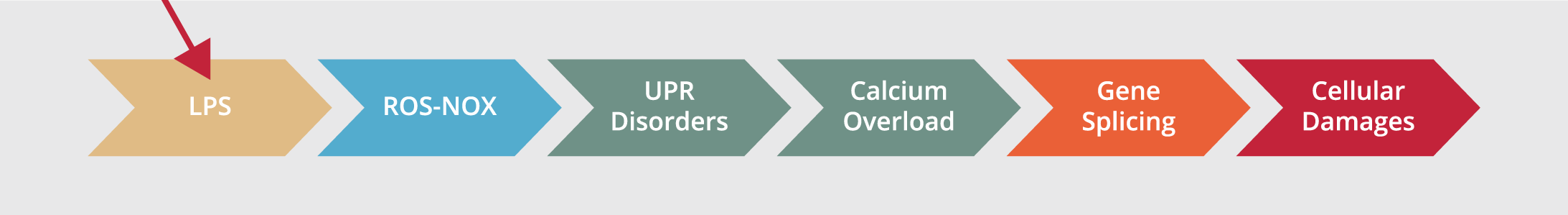
Figure 1. Study hypothesis over LUCOGS.
There are different inflammatory and antioxidant mechanisms available in the hepatic cell during exposure to LPS. Ca+2 released by inositol three phosphate receptor (IP3R) increases mitochondrial activity during endoplasmic reticulum (ER) stress to revert energy imbalance, but if it fails, the cell will die as a result of the mitochondrial apoptotic cascade. Ca+2 released from ryanodine receptors on the endoplasmic reticulum could increase the Ca+2 overload pool for IP-3 receptors and lead to an extra inflammatory response.
ROS: Reactive Oxygen Species, UPR: Unfolded Protein Response, LPS: Lipopolysaccharide, NOX: NO and NO2, ROS: Reactive Oxygen Species.
In our previous research study, we showed that hLPS could increase cytokine release (8). However, in the present study, we investigated hLPS effects on LUCOGS and focused on the UPR machinery and correlated signal transduction inducing gene splicing in mice’s hepatic tissue (Figure 1).
Quality Control and Assurance of Human LPS Isolated from Burned Wound
The isolated bacteria from human burned wounds were confirmed as P. aeruginosa sort, based on culture and biochemical identification observations and a positive oxidase test (Figure 2, A-H). The PAGE electrophoresis data show a different concentration of purified hLPS. The same samples with an efficient concentration were pooled together and applied for the next steps of the tests.
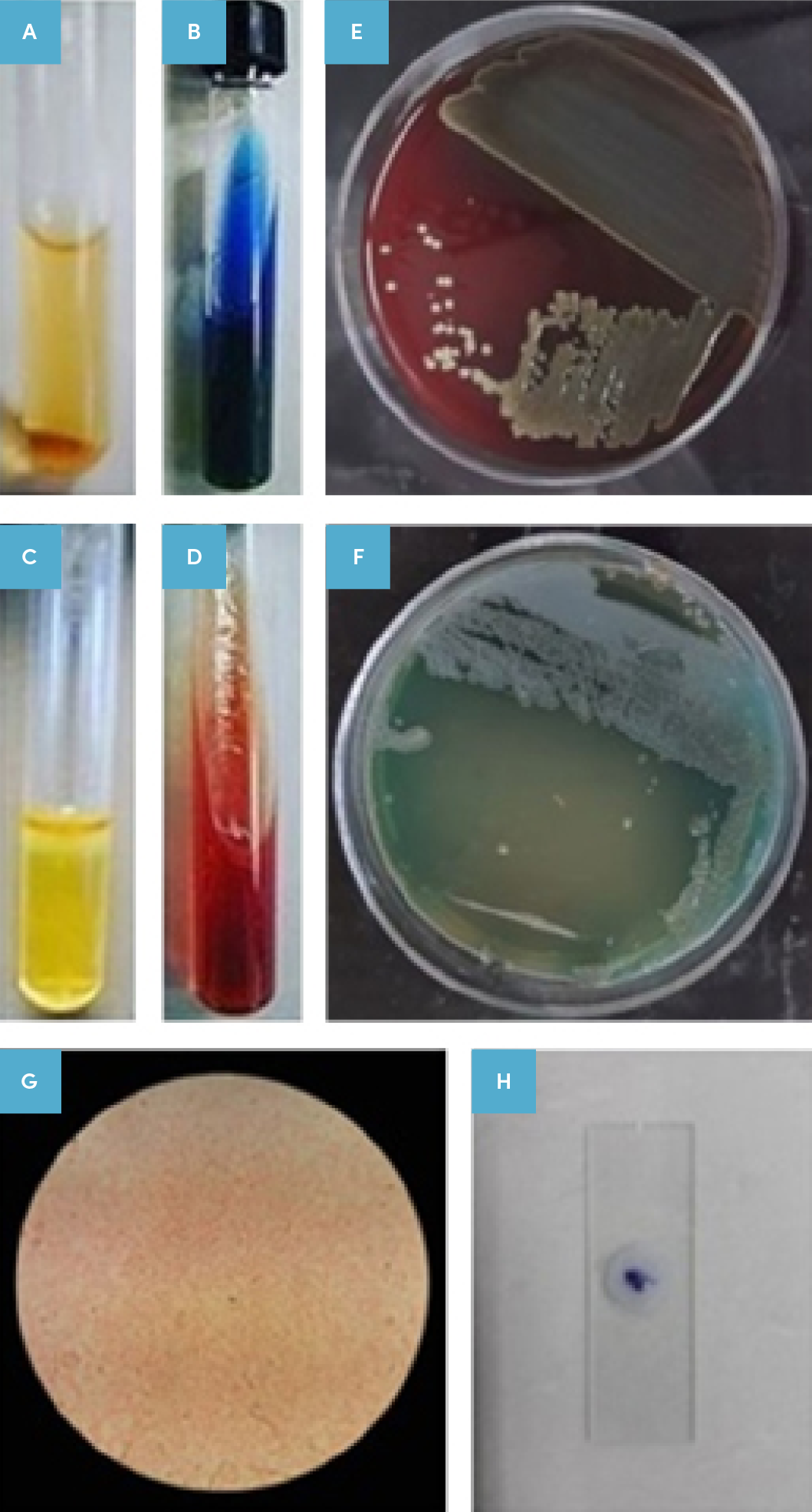
Figure 2. Isolated bacterium determination.
Results of the isolated bacteria identification of patients based on culture, microscopic examination, and biochemical test results.
A: Urease negative test, B: Positive Simon citrate test, C: Sulfur indole motility (SIM) test, D: Triple sugar iron agar (TSI) test (alkaline/alkaline), E: Positive cultivation on Blood Agar medium, F: Positive cultivation and production of specific pigment in cetrimide agar,
G: Gram negative staining in the microscopic examination; and
H: Positive oxidase test.
In addition, Figure 2 depicts how we isolated targeted bacteria and hLPS from a human wound. In short, after obtaining informed consent from the patients, burned wound bacteria were isolated and cultured on the blood agar. Cultured bacteria from specimens grown well in enriched, specific, and differential media (Figure 2, A-H).
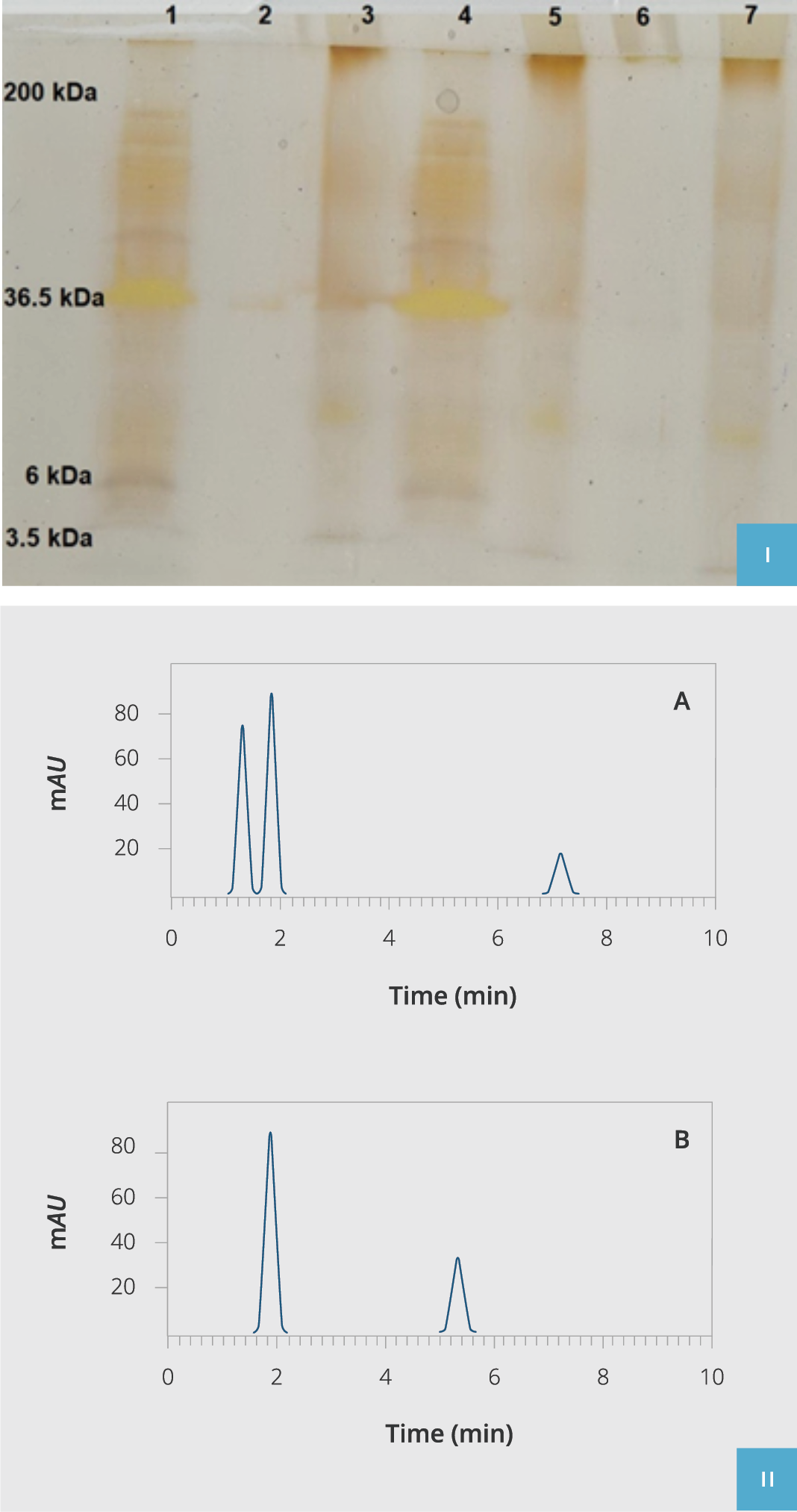
Figure 3. Extracted hLPS validation.
I) hLPS poly acrylamide gel electrophoresis (PAGE). Lane 1: Molecular weight ladder (3.5-200kDa), Lanes 2, 3, 5, and 7: Purified hLPS of P. aeruginosa isolated from clinical samples, Lane 4: Standard commercial control of P. aeruginosa LPS (SIGMA-Cat: L8643).
II) HPLC fractionation of purified hLPS compared to the standard commercial control of P. aeruginosa LPS (SIGMA-Cat: L8643). Chromatogram A indicates purified hLPS from isolated clinical samples, and chromatogram B is the standard commercial control of P. aeruginosa LPS (SIGMA-Cat: L8643). HPLC: High-Performance Liquid Chromatography, hLPS: The LPS extracted from P. aeruginosa.
After isolation and validation of human targeted bacteria sort via biochemical tests, the hLPS biologic compound was purified, and its quality and efficacy were compared to standard chemical LPS using HPLC (Figure 3, A and B). We observed that our isolated purified hLPS had proper purity as standard LPS checked by 12% PAGE silver staining. Eventually, all hLPS samples were aliquoted and stocked at -70°C for further supplementary tests (final concentration 3mg/ml).
Molecular Analysis
The molecular results acquired from whole experiments after confirmation of splicing and non-splicing pattern were represented as follows:
Two-hour post-injection
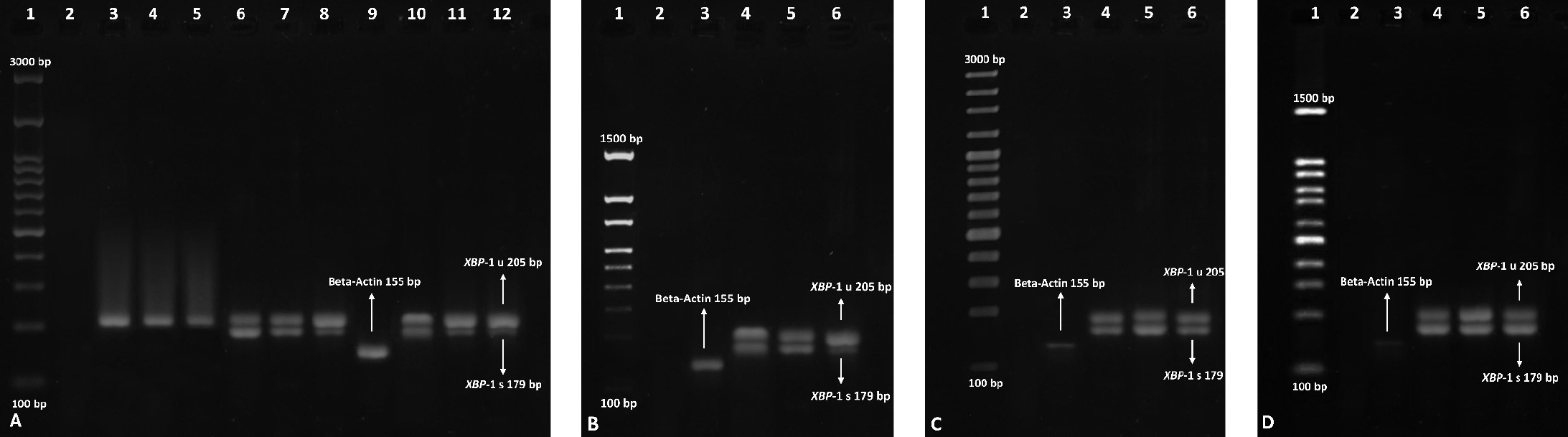
Figure 4. Monitoring the splicing of XBP-1 in the liver.
A) Controls. Left to right: 1) Molecular weight DNA ladder (100-3000 bp), 2) RT-PCR negative control of RT-PCR, 3-5) Mice negative control groups, 6-8) Mice positive control group using an UPR activating inducer (tunicamycin), 9) RT-PCR positive internal control by beta-actin housekeeping gene amplification (155bp), 10-12) Mice positive controls treated with hLPS.
B) Mice treated with hLPS+CCI1. Left to right: 1) Molecular weight DNA ladder (100-1500 bp), 2) RT-PCR negative control, 3) RT-PCR positive internal control by beta-actin housekeeping gene amplification (155bp), 4-6) Mice treated with hLPS+CCI1.
C) Mice treated with hLPS+CCI2. Left to right: 1) Molecular weight DNA ladder (100-3000 bp), 2) RT-PCR negative control, 3) RT-PCR positive internal control by beta-actin housekeeping gene amplification (155bp), 4-6) Mice treated with hLPS+CCI2
D) Mice treated with hLPS+CCI1+CCI2. Left to right: 1) Molecular weight DNA ladder (100-1500 bp), 2) RT-PCR negative control, 3) RT-PCR positive internal control by beta-actin housekeeping gene amplification (155bp), 4-6) Mice treated with hLPS+CCI1+CCI2.
The whole treatment was assessed for the three-time interval for all mice (2, 8, and 24 hours after injections). RT-PCR: Reverse-Transcriptase Polymerase Chain Reaction.
In the negative control group, none of the animals exhibited any XBP-1 splicing (Figure 4, A-D). We observed that all six mice showed XBP-1 splicing in the first positive control group (receiving tunicamycin, 2.5 mg/kg), and three out of six mice in the second positive control group (hLPS, 3mg/kg) 2 hours after the stimulation (Figure 4, A). In mice receiving the combination of hLPS+CCI1, four out of six mice showed splicing (Figure 4, B), but three out of six mice showed XBP-1 splicing in the group, which received hLPS+CCI2 (Figure 4, C). Surprisingly, all six out of six mice showed an increase in the XBP-1 splicing after injecting both CCIs (hLPS+CCI1+CCI2) at the 2nd hour (Figure 4, D).
Eight-hour post-injection
In the negative control group, none of the animals showed any XBP-1 splicing (Figure 4, A). We observed that all six mice showed XBP-1 splicing in the first positive control group (receiving tunicamycin, 2.5 mg/kg) and two out of six mice in the second positive control group (hLPS, 3mg/kg) 8 hours after the injection. Four out of six mice that received the combination of hLPS+CCI1, and three out of six mice that received hLPS+CCI2, exhibited XBP-1 splicing. Surprisingly, all six mice showed an increase in the XBP-1 splicing after injecting both CCIs (hLPS+CCI1+CCI2) for 8 hours stimulations the same as 2 hours (Figure 4, D). It was a statistically significant difference compared to the hLPS group (p = 0.03).
Twenty-four-hour post-injection
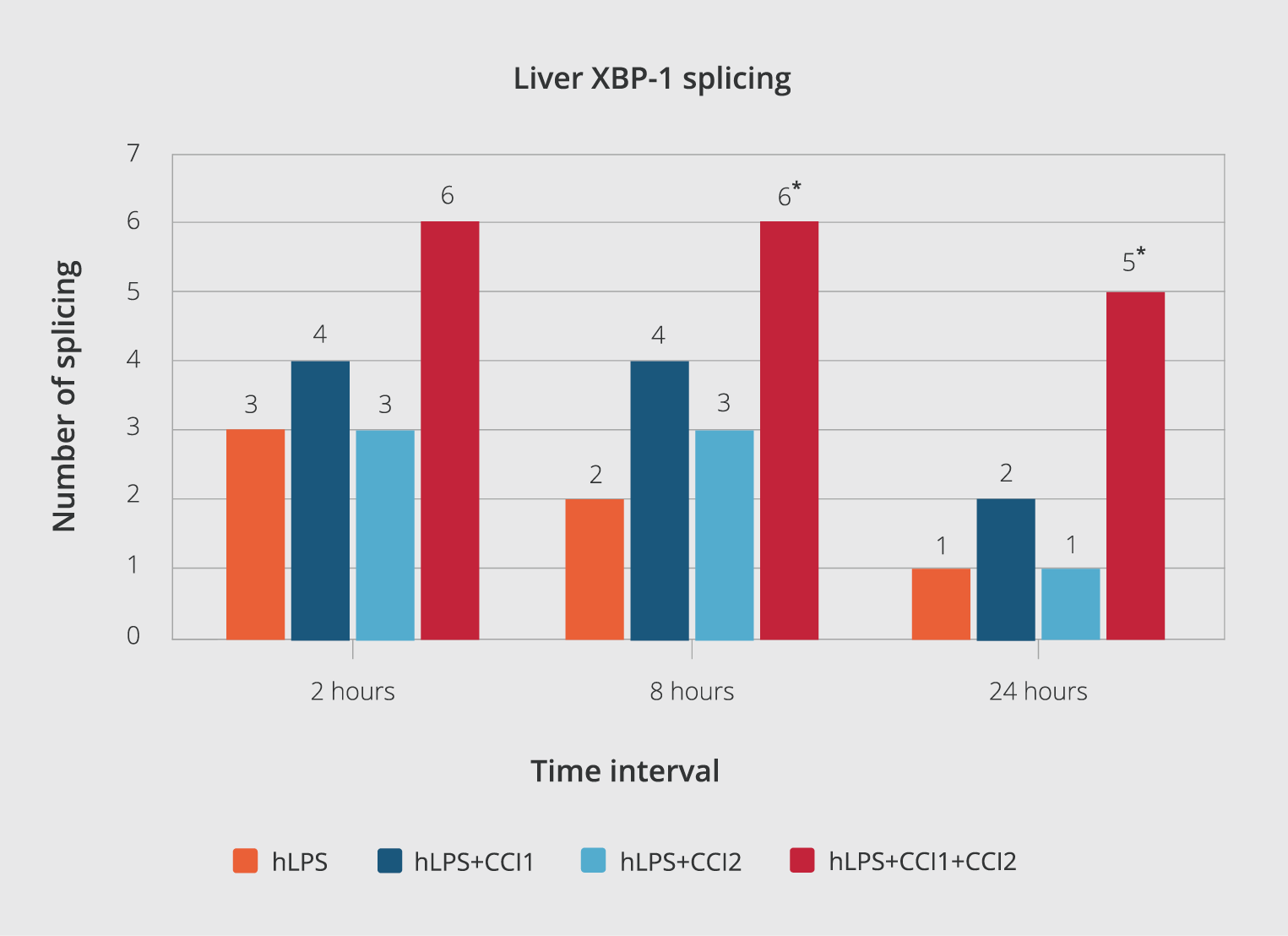
Figure 5. The number of spliced XBP-1 at different intervals.
XBP-1 splicing results were reported at three-time intervals at 2, 8, and 24 hours after each injection. The results of the tests were analyzed via the χ2 statistic test (significant level p< 0.05%). All data have been expressed as mean ± standard deviation. (*) indicate p< 0.05 compared to the hLPS group control.
In the negative control group, none of the animals exhibited any XBP-1 splicing (Figure 4, A). We observed that all six mice showed XBP-1 splicing in the first positive control (receiving tunicamycin, 2.5 mg/kg). Only one out of six mice in the second positive control (hLPS, 3mg/kg) showed XBP-1 splicing after 24 hours of stimulation. In mice receiving the combination of hLPS+CCI1, two out of six mice showed splicing 24 hours after the injection (Figure 4, B). In the hLPS+CCI2 group, only one out of the six mice showed XBP-1 splicing (Figure 4, C). Unpredictably, in the CCIs (hLPS+CCI1+CCI2) group, five out of six mice showed XBP-1 splicing (Figure 4, D). There was a significant statistical difference in the number of XBP-1 splicing and XBP-1s production only in the group that received both CCI1 and CCI 2 than in the hLPS group (Figure 5).
DISCUSSION
The important novelty of this study is the use of natural endotoxin purified from bacteria isolated from infected burn wounds of patients. We reconstructed this isolated toxin in an animal in vivo model system to observe pathological effects of LPS clearly. Previously, as another study group also described, we demonstrated that LPS had cytotoxic effects on increasing cytokine release from different tissues and cells (8). In this study, we show that our extracted hLPS from P. aeruginosa can affect pro-inflammatory XBP-1 related gene splicing. XBP-1 splicing correlates with the UPR activation and calcium overload through cells, and leads to random organ shutdown.
Many studies have been conducted on the interactions between the Ca2+ ion hemostasis and the UPR machinery, and how Gram-negative bacteria’s LPS induces septic shock (1, 2). Nonetheless, there are limited studies, which used purified wild LPS from a human burn wound (which we called hLPS in this study) and/or from the wild colonized bacteria at the site of the wound’s infection.
After describing our goals and main objectives, the Medical Science Committee allowed us to study different aspects of organ shutdown mechanisms using animal modeling as a representative in vivo model system. In the traditional animal model of cancer, limitations in representing features of each cancer patient led to established patient-derived xenograft (PDX) animal model systems (44), which helped medicine to develop appropriate drugs. With the same concept, we consider building a micro-environment of wild bacteria (endotoxins), which is colonized in the patient’s burn wound. Subsequently, we attempt to extract wild LPS instead of applying commercial LPS to establish a new animal model of endotoxemia with more ‘natural stimulators’ (hLPS vs. commercial LPS) that might better represent pathophysiologic conditions. We assumed that this novel animal model would represent a more accurate model system and could yield real and natural results after experiments.
After establishing the animal model, we focused on our observed data according to treatment with our target CCIs formulae. The more XBP-1 splicing and XBP-1s production are, the more anti-inflammatory event may occur. Consecutively, the subject’s biologic system can equip better itself to respond to endotoxemia appropriately.
Hypothetically, the stress in the eukaryotic endoplasmic reticulum results in the activation of UPR in response to increased levels of miss-folded proteins and maintenance of homeostasis caused by endotoxin-induced P. aeruginosa’s LPS, which plays a key role in this pathway.
Furthermore, Ca+2 ions, compared to other ions, play an essential role in the homeostasis of eukaryotic cells (18-22).
Our study results showed that infection with P. aeruginosa can be used to determine the destiny of cells or tissues by encountering one of its major pathogens secretomes, LPS. This effect will often indicate its presence, in the form of increasing the bioavailability of the living tissue and cell or causing the death of the cell and ultimately the subject (23).
Pierobon N, et al. (33) postulated that the second messenger IP3 elicited Ca2+ signals control many important processes in hepatocytes, including mitochondrial metabolism, glycogen degradation and gene expression. This signal is modulated in response to various molecules that mainly act through the well-known phosphoinositide cascade. The essential components of this signaling pathway are the formation of IP3 at the plasma membrane, binding of IP3 to IP3Rs, and release of Ca2+ into the intracellular space of cells.
The use of CCIs in this study showed a significant increase in the splicing of XBP-1 mRNA, and in the production of sXBP-1 transcriptomes, which are important to control pro-inflammatory cytokines. This increment leads to a beneficial effect on decreasing the amount of inflammatory mediators, which play determining roles in dealing with septic shock and subsequently increase (pro-)inflammatory side effects (10, 15, 20).
Many bacterial virulence factors play crucial roles in inflammation and septic shock. One of these toxic factors is LPS, which after absorption from different pathways, can cause (pro-)inflammatory cytokine release, such as IL-1b, IL-7, IL-6, and TNF-α (45). Depending on the target tissue, these (pro-)inflammatory mediators can determine the severity and fate of the affected tissue. Usually, these mediators are affected by cell metabolism through direct and indirect processes.
In the direct pathway, bacterial LPS is attached to receptors, such as TLR-4 and CD-14. Afterward, nuclear cell signaling leads to product inflammatory mediators, such as IL- 1, IL-6, and TNFα (25).
In the indirect pathway, the activation of the related pathway is accomplished by absorbing LPS in a path other than the TLR-4 receptor, which can lead to cell death (26). To cause cell death, calcium overload of cells occurs in different ways (17-19, 21-23). One of the most important entering routes for these ions is Ca+2 channels. The entrance of Ca+2 ions occurs coincidentally in cellular inflammations resulting from LPS (19-21). The two most important cellular channels in the excessive intake of Ca+2 that lead to cell death are ryanodine and IP3 channel (21). However, there is no clear evidence for expression or a role of ryanodine receptors (RyRs) in the liver, but some studies indirectly suggest its involvement. Furthermore, there is no report about therapeutic and anti-inflammatory effects of CCIs for the endotoxic shock caused by P. aeruginosa’s LPS (20). P. aeruginosa is considered an important opportunistic pathogen in nosocomial infections of burn wounds (4, 20, 46), and it might be a colonizer in different trauma patients. The results of treatment with combination of both CCI1 and CCI2 in our study, which are inhibitors of the ryanodine and IP3 channel, suggest that treating with two different CCIs exerts much more appropriate inhibitory effects than treatment with separate CCIs does (Figure 5). This additive/synergistic effect directly correlates with the amount of XBP-1 gene transcription factor mRNA splicing. The CCIs prescription can activate adaptive UPR’s signals by inducing XBP–1 splicing and producing XBP-1s, and it can play a crucial role in the maintenance of hepatic cells. It is highly important to save liver vital capacity owing to its significance as a headquarter of the detoxification organ in the human body (47).
It appears that the more Ca2+ channels become inhibited, the more adaptive UPR gene-splicing levels increase subsequently. As described previously (39) and confirmed with our results, this opportunity could be enhanced further by increasing the activity of the IRE-1’s sensor and activating XBP-1 spliced products.
Based on the findings of our study, it can be stated that the combination of two types of CCIs has a higher potency to prevent the entry of considerable amounts of Ca+2 ions into the cell in 8 or 24 hours after induced-endotoxemia with hLPS. The inhibitory effect of using different CCIs in combination is stronger than the use of one type of CCIs separately.
As to conclude, we showed that in the near future, it could be possible to study the LUCOGS system in vivo and investigate different aspects of the signal transductions correlated with (un-)known pro-inflammatory-endotoxemic-UPR-gene-splicing processes by using our human-animal model system. Moreover, appropriate synergistic combination drugs of CCIs may prevent SRPs in patients, requiring future investigations. For future studies to clarify more mechanistic details about functions of CCIs to develop different anti-septic drugs, we strongly recommend conducting more evaluations about UPR biosensors like PERK and ATF-6 qualitatively and quantitatively, i.e., investigating a certain protein expression and its related gene activity associated changes.
Informed Consent
Written informed consents for publication of their clinical details were obtained from the patients.
Ethical Approval
The Guilan Medical Committee approved the study on October 23, 2017, with the decision number IR.GUMS.REC.1396.81.
Peer-review
Externally peer-reviewed
Author Contributions
Concept – M.H.C., M.A.; Design– M.H.C., M.M.; Supervision – M.H.C., M.S., M.M., M.A., A.N., A.C.K.; Fundings – M.H.C., M.M.; Materials – M.S., A.N.; Data Collection and/or Processing – M.S., A.C.K.; Analysis and/or Interpretation – M.A.; Literature Review– A.C.K., A.N.; Writer – M.H.C., M.S., M.M., M.A., A.N., A.C.K.; Critical Review – M.H.C., M.S., M.M., M.A., A.N., A.C.K.
Conflict of Interest
The authors have no conflict of interest to declare.
Financial Disclosure
This project has been approved as a research project with the financial support of the vice-chancellor of Research and Technology, Guilan University of Medical Sciences with the Research Code 96023010.
Acknowledgments
The authors thank Mr. Morrovati H, Animal Lab, Guilan University, Faculty of Medicine for his technical assistance.
References
- Munford RS, Varley AW. Shield as signal: lipopolysaccharides and the evolution of immunity to gram-negative bacteria. PLoS Pathog. 2006;2(6):e67. [CrossRef]
- McCormick JK, Yarwood JM, Schlievert PM. Toxic shock syndrome and bacterial superantigens: an update. Annu Rev Microbiol. 2001;55:77-104. [CrossRef]
- Morrison DC, Ulevitch RJ. The effects of bacterial endotoxins on host mediation systems. A review. Am J Pathol. 1978;93(2):526-618.
- Alexander C, Rietschel ET. Bacterial lipopolysaccharides and innate immunity. J Endotoxin Res. 2001;7(3):167-202.
- Rudd KE, Kissoon N, Limmathurotsakul D, et al. The global burden of sepsis: barriers and potential solutions. Crit Care. 2018;22(1):232. [CrossRef]
- Van Amersfoort ES, Van Berkel TJ, Kuiper J. Receptors, mediators, and mechanisms involved in bacterial sepsis and septic shock. Clin Microbiol Rev. 2003;16(3):379-414. [CrossRef]
- Maldonado RF, Sa-Correia I, Valvano MA. Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiol Rev. 2016;40(4):480-93. [CrossRef]
- Badlou BA. Combination of calcium channels inhibitors prevents LPS – Induced (pro-) inflammatory release of the TNF-α and IL-1β, in vivo”. EC Cardiology 7.3. 2020;01-09.
- Grootjans J, Kaser A, Kaufman RJ, Blumberg RS. The unfolded protein response in immunity and inflammation. Nat Rev Immunol. 2016;16(8):469-84. [CrossRef]
- Janssens S, Pulendran B, Lambrecht BN. Emerging functions of the unfolded protein response in immunity. Nat Immunol. 2014;15(10):910-9. [CrossRef]
- Niu M, Dai X, Zou W, et al. Autophagy, endoplasmic reticulum stress and the unfolded protein response in intracerebral hemorrhage. Transl Neurosci. 2017;8:37-48. [CrossRef]
- Pillich H, Loose M, Zimmer KP, Chakraborty T. Diverse roles of endoplasmic reticulum stress sensors in bacterial infection. Mol Cell Pediatr. 2016;3(1):9. [CrossRef]
- Hetz C, Martinon F, Rodriguez D, Glimcher LH. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol Rev. 2011;91(4):1219-43. [CrossRef]
- Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454(7203):455-62. [CrossRef]
- Santafe MM, Urbano FJ, Lanuza MA, Uchitel OD. Multiple types of calcium channels mediate transmitter release during functional recovery of botulinum toxin type A-poisoned mouse motor nerve terminals. Neuroscience. 2000;95(1):227-34. [CrossRef]
- Shinde V, Kotla P, Strang C, Gorbatyuk M. Unfolded protein response-induced dysregulation of calcium homeostasis promotes retinal degeneration in rat models of autosomal dominant retinitis pigmentosa. Cell Death Dis. 2016;7:e2085. [CrossRef]
- Atlas D. Functional and physical coupling of voltage-sensitive calcium channels with exocytotic proteins: ramifications for the secretion mechanism. J Neurochem. 2001;77(4):972-85. [CrossRef]
- Berridge MJ. Calcium signalling remodelling and disease. Biochem Soc Trans. 2012;40(2):297-309. [CrossRef]
- Kandasamy K, Bezavada L, Escue RB, Parthasarathi K. Lipopolysaccharide induces endoplasmic store Ca2+-dependent inflammatory responses in lung microvessels. PLoS One. 2013;8(5):e63465. [CrossRef]
- Mikoshiba K. Role of IP3 receptor signaling in cell functions and diseases. Adv Biol Regul. 2015;57:217-27. [CrossRef]
- Bahar E, Kim H, Yoon H. ER stress-mediated signaling: Action potential and Ca(2+) as key players. Int J Mol Sci. 2016;17(9):1558. [CrossRef]
- Hajnoczky G, Robb-Gaspers LD, Seitz MB, Thomas AP. Decoding of cytosolic calcium oscillations in the mitochondria. Cell. 1995;82(3):415-24. [CrossRef]
- Han DW. Intestinal endotoxemia as a pathogenetic mechanism in liver failure. World J Gastroenterol. 2002;8(6):961-5. [CrossRef]
- Lin RS, Lee FY, Lee SD, et al. Endotoxemia in patients with chronic liver diseases: relationship to severity of liver diseases, presence of esophageal varices, and hyperdynamic circulation. J Hepatol. 1995;22(2):165-72. [CrossRef]
- Zughaier SM, Zimmer SM, Datta A, Carlson RW, Stephens DS. Differential induction of the toll-like receptor 4-MyD88-dependent and -independent signaling pathways by endotoxins. Infect Immun. 2005;73(5):2940-50. [CrossRef]
- Pugin J, Ulevitch RJ, Tobias PS. Activation of endothelial cells by endotoxin: direct versus indirect pathways and the role of CD14. Prog Clin Biol Res.1995;392:369-73.
- Wang Z, Zhou Y, Yu Y, He K, Cheng LM. Lipopolysaccharide preconditioning increased the level of regulatory B cells in the spleen after acute ischaemia/reperfusion in mice. Brain Res. 2018;1701:46-57. [CrossRef]
- Delcour AH. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta. 2009;1794(5):808-16. [CrossRef]
- Farrag HA, Abdallah N, Shehata MMK, Awad EM. Natural outer membrane permeabilizers boost antibiotic action against irradiated resistant bacteria. J Biomed Sci. 2019;26(1):69. [CrossRef]
- Pruitt BA, Jr., McManus AT, Kim SH, Goodwin CW. Burn wound infections: current status. World J Surg. 1998;22(2):135-45. [CrossRef]
- Hakemi MS. Chronic kidney disease epidemiology. Iran J Kidney Dis. 2014;8(4):261-2.
- Rahme LG, Tan MW, Le L, et al. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc Natl Acad Sci U S A. 1997;94(24):13245-50. [CrossRef]
- Pierobon N, Renard-Rooney DC, Gaspers LD, Thomas AP. Ryanodine receptors in liver. J Biol Chem. 2006;281(45):34086-95. [CrossRef]
- Pedersen SS. Lung infection with alginate-producing, mucoid Pseudomonas aeruginosa in cystic fibrosis. APMIS Suppl. 1992;28:1-79.
- Pier GB. Pseudomonas aeruginosa lipopolysaccharide: a major virulence factor, initiator of inflammation and target for effective immunity. Int J Med Microbiol. 2007;297(5):277-95. [CrossRef]
- Dolmetsch RE, Xu K, Lewis RS. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392(6679):933-6. [CrossRef]
- Duan K, Dammel C, Stein J, Rabin H, Surette MG. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol Microbiol. 2003;50(5):1477-91. [CrossRef]
- Pirnay JP, De Vos D, Cochez C, et al. Pseudomonas aeruginosa displays an epidemic population structure. Environ Microbiol. 2002;4(12):898-911. [CrossRef]
- Back SH, Lee K, Vink E, Kaufman RJ. Cytoplasmic IRE1alpha-mediated XBP1 mRNA splicing in the absence of nuclear processing and endoplasmic reticulum stress. J Biol Chem. 2006;281(27):18691-706. [CrossRef]
- Chalmers F, van Lith M, Sweeney B, Cain K, Bulleid NJ. Inhibition of IRE1alpha-mediated XBP1 mRNA cleavage by XBP1 reveals a novel regulatory process during the unfolded protein response. Wellcome Open Res. 2017;2:36. [CrossRef]
- Remondelli P, Renna M. The endoplasmic reticulum unfolded protein response in neurodegenerative disorders and its potential therapeutic significance. Front Mol Neurosci. 2017;10:187. [CrossRef]
- Nemzek JA, Hugunin KM, Opp MR. Modeling sepsis in the laboratory: merging sound science with animal well-being. Comp Med. 2008;58(2):120-8.
- Samali A, Fitzgerald U, Deegan S, Gupta S. Methods for monitoring endoplasmic reticulum stress and the unfolded protein response. Int J Cell Biol. 2010;2010:830307. [CrossRef]
- Jung J, Seol HS, Chang S. The generation and application of patient-derived xenograft model for cancer research. Cancer Res Treat. 2018;50(1):1-10. [CrossRef]
- Assimacopoulos-Jeannet FD, Blackmore PF, Exton JH. Studies on alpha-adrenergic activation of hepatic glucose output. Studies on role of calcium in alpha-adrenergic activation of phosphorylase. J Biol Chem. 1977;252(8):2662-9.
- Özer-Balın Ş, Aktaş-Şenol A. [Evaluation of hospital infections in intensive care unit]. Klimik Derg. 2017;30(3):108-13. Turkish. [CrossRef]
- Huang N, Yu Y, Qiao J. Dual role for the unfolded protein response in the ovary: adaption and apoptosis. Protein Cell. 2017;8(1):14-24. [CrossRef]